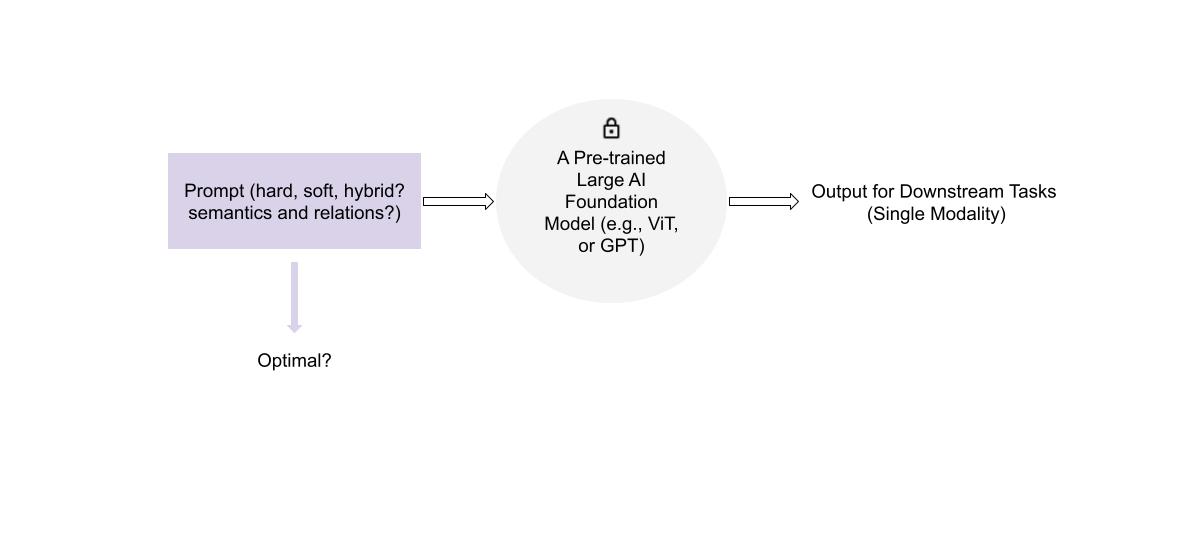
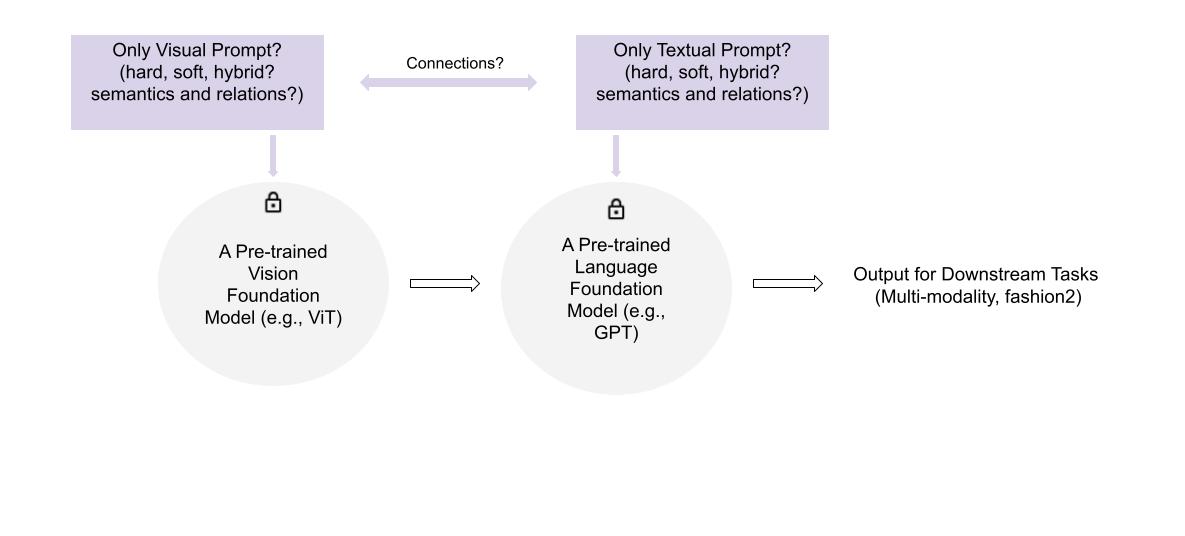
Visual Prompt Tuning for AI Foundation Models
Abstract: Large Pre-trained AI foundation models have great potentials to be leveraged in various downstream tasks. However, directly
fine-tuning them in downstream domains confronts challenges, including limited data volume, severe domain gap, and costly
training expense. While learnable prompts can be used to tune a foundation model, it remains unclear what kinds of prompts are
mostly beneficial to specific tasks. This research aims to investigate designing optimal prompts for downstream tasks, and to explore
appropriate ways for learning such prompts.
Semi-supervised Deep Active Learning
Abstract: Most classical active learning algorithms only use labeled data to train a task model. Our investigation shows that ample unlabeled data can
arguably provide rich and useful information for the task model training. In this research, we aim to design novel schemes to properly involve
unlabeled data in training the task model, while the schemes are also expected to serve as new metrics for estimating the uncertainty of
unlabeled data.
Interpretable Active Learning
Abstract: The ultimate goal of active learning is to train a powerful task model with a certain number of newly annotated data. The capability of the
task model should be evaluated by its testing performance. Thus, a question can be naturally raised: how an individual annotated data sample will
impact the final testing performance. In this research, we aim to investigate and interpret such an impact, which can inversely provide a guidance
for unlabeled data selection in active learning.
Cold-start Active Learning
Abstract: Cold-start active learning means there is no initial labeled data that can be adopted to train a task model, and an AL algorithm is expected to
select the most uncertain or representative data samples based on very little prior knowledge. Such a circumstance is very common in biomedical
research. In this research, we aim to develop reliable cold-start active learning pipelines for unlabeled data selection, reducing annotation cost
for biomedical researchers.
Relations between Machine Learning Fashions
Abstract: In this research, we aim to explore the relations between various machine learning fashions. For instance, how active learning relates to
curriculum learning, and whether one can improve the other. We expect such a study to foster a deep understanding of various learning fashions,
eventually benefiting the development of robust machine learning pipelines.
Data Aspect AI
Abstract: Most existing AI research focuses on model aspect (e.g. designing novel networks or training schemes). Data aspect AI is an emerging and promising
research area, gaining tremendous attention from both academia and industry. In this research, we aim to propose and leverage novel data aspect AI
solutions to boost biomedical data analytics.
Automated Science
Abstract: Automated science is an emerging brand-new research area. In this research, we aim to develop a reliable platform to help biologists optimize
their experimental designs. The core of automated science is active machine learning, and we leverage our advantages on active learning research
to explore this new area.
Cryo-electron Tomography (Cryo-ET) Analysis with Machine Learning
Abstract: Cryo-Electron Tomography (cryo-ET) is an emerging 3D imaging technique which shows great potentials in structural biology research.
In this research, we aim to develop novel machine learning algorithms to address the challenges in Cryo-ET analysis, such as semantic
segmentation and denoising. The following images are cited from XuLab.